IUCN/SSC Otter Specialist Group Bulletin

©IUCN/SCC Otter Specialist Group
Citation: De Ferran, V., Chua, C.E.E., Johns, P., Eizirik, E, and Koepfli, K.-P. (2025). Large-Scale Identification of Microsatellite Loci from Multiple Otter (Mammalia, Carnivora, Lutrinae) Species using Whole Genome Sequence Data. IUCN Otter Spec. Group Bull. 42 (3): 120 - 132
Large-Scale Identification of Microsatellite Loci from Multiple Otter (Mammalia, Carnivora, Lutrinae) Species using Whole Genome Sequence Data
Vera De Ferran1*,Channelle Ee Eun Chua2, Philip Johns2, Eduardo Eizirik1,3, and Klaus-Peter Koepfli4*
1UPontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Escola de Ciências da Saúde e da Vida, Laboratório de Biologia Genômica e Molecular, Porto Alegre, RS, Brazil
2Yale-NUS College, 16 College Ave West, Singapore
3Instituto Pró-Carnívoros, Atibaia, São Paulo, Brazil
4Smithsonian-Mason School of Conservation, George Mason University, Front Royal, VA, 22630, USA
*Corresponding Author Email: veradeferran@gmail.com and klauspeter.koepfli527@gmail.com
Received 11th April 2025, accepted 27th April 2025
Abstract: The development of molecular studies on elusive, rare, and/or poorly known species faces challenges due to the lack of suitable markers. Species-specific microsatellite markers minimize bias, offer better performance, and are cost-effective, aiding the development of population genetic studies. The use of whole-genome sequences allows for the development of species-specific microsatellite markers and their survey in closely related species, enabling the discovery of shared markers that can facilitate comparative studies. Lutrinae includes 14 extant species of otters. Despite their worrisome conservation status, due to inherent characteristics of these species that make their study difficult by traditional methods, many of them lack reliable population genetic data, limiting conservation efforts. In this study, we employed a multi-taxon approach to identify a large number of novel microsatellite loci for 11 of the 14 otter species, assessing whether the identified loci were shared among different taxa. We identified a total of 23,320 microsatellite loci across 11 species, which were reduced to 12,573 after stringent filtering criteria. Primer design was completed successfully for 420 and 259 unique loci, considering two minimum melting temperatures. We validated marker efficiency by testing the 81 loci designed for two Asian species. Of these, 51 loci yielded reliable microsatellite genotypes in both species, with 34 showing allelic variation in at least one of them. These results demonstrate that these markers are applicable in empirical genotyping for both their target species and closely related ones.
Keywords: Otters, microsatellites, genome sequences, conservation genetics, genetic monitoring
INTRODUCTION
Molecular approaches have revolutionized the fields of genetics, ecology, and evolutionary biology, and are routinely applied in surveys, monitoring programs and status assessments of threatened organisms (Schwartz et al., 2007). However, the application of such methods to wildlife species that are rare, elusive, and/or poorly studied is often hindered by the paucity of appropriate molecular markers for efficient genetic analysis. Therefore, developing molecular tools that are specifically designed for poorly known taxa is a useful avenue to enable genetic, ecological, and conservation-oriented studies targeting these species.
Of the various molecular markers developed in the last four decades to survey natural populations, microsatellite loci have been shown to be particularly informative for multiple types of problems (Frankham et al., 2002). For example, they have been used in many different taxa to assess levels of genetic diversity, population structure and demographic connectivity, as well as to perform individual-based ecological, behavioral, and forensic analyses (e.g., Trinca et al., 2013; Miller et al., 2014; Ishida et al., 2018; Harper et al., 2018; Sun et al., 2020). Despite their usefulness, the routine application of microsatellites to address such problems in wildlife species has been historically hampered by the lack of markers developed specifically for the target organism, often requiring their transfer (i.e., use of the same primers with adjusted PCR conditions) from better-known, related species (Frankham et al., 2002). Although this approach is often successful, species-specific markers are expected to show better performance in terms of variability (informative content) and PCR efficiency (which allows, for example, improved recovery of molecular data from degraded field-collected materials such as feces and hairs). In addition, their use should minimize ascertainment bias associated with using heterologous loci (Li and Kimmel, 2013).
Despite the increasing application of reduced representation and low-coverage whole genome sequencing approaches to analyze single nucleotide polymorphisms (Fuentes-Pardo and Ruzzante, 2017), microsatellites remain a cost-effective and viable tool for population genetic analyses of animal and plant populations, especially in species with reduced genetic diversity (Hauser et al., 2021) or that are being genetically surveyed for the first time. Genome sequencing has greatly facilitated the development of species-specific microsatellite markers by enabling the rapid computational discovery of repetitive sequences from genomic data (Benson, 1999). Although this strategy has now been used for many systems (e.g., Kumari et al., 2019; Liu et al., 2019; Latorre-Cardenas et al., 2020), and it is starting to become more common with the increase in the availability of genomic resources, it is still not commonly applied to whole-genome sequences from multiple species of the same clade (but see Liu et al., 2017; Xu et al., 2018; Bhat et al., 2018; Manee et al., 2020 for examples). Such an approach would allow the parallel survey of microsatellites in closely related species, enabling the discovery of shared markers that can facilitate comparative studies.
An interesting clade in which this approach can be assessed is the mustelid subfamily Lutrinae, which comprises 14 extant species of otters. The current IUCN Red List of Threatened Species (IUCN, 2022) categorizes otter species as: ‘Least Concern’ (Lontra canadensis, North America); ‘Near Threatened’ (Aonyx capensis, Sub-Saharan Africa; Aonyx congicus, Central Africa; Hydrictis maculicollis, Sub-Saharan Africa; Lontra longicaudis, South America; Lutra lutra, Eurasia and North Africa); ‘Vulnerable’ (Aonyx cinereus, Lutrogale perspicillata, South and Southeast Asia); and ‘Endangered’ (Enhydra lutris, North Pacific Rim; Lontra felina, Pacific coast of South America; Lontra provocax, southern South America; Lutra sumatrana, Southeast Asia; Pteronura brasiliensis, South America). The recently recognized Lontra annectens of Central America (de Ferran et al., 2024) has not been assessed yet by the IUCN. Despite their worrisome conservation status and the estimated reduction in census sizes for most of these species, there is no reliable data on demographic, ecological and genetic aspects for most of these taxa throughout most of their respective geographic distributions (Enhydra lutris being a notable exception, e.g., Larson et al., 2021; Beichman et al., 2023), which limits conservation planning and action.
Otters have several characteristics that make their study difficult by traditional methods: most species (except Pteronura brasiliensis and Hydrictis maculicollis) do not present a coat pattern (e.g., spots or stripes) that allow individual identification; they are semi-aquatic, limiting the use of camera-traps or radio-telemetry methods and also making it difficult to capture and immobilize these animals; and several species are secretive and not commonly observed in the wild, in addition to being crepuscular/nocturnal. For these reasons, most studies on these species are based on indirect records, such as using feces and latrines as sources of information on presence, diet, home range size, and activity (e.g., Crowley et al., 2012; Rivera et al., 2019). However, these methods are quite limited for population studies due to the impossibility of identifying individuals with morphology-based methods, along with the difficulty in differentiating species in areas where more than one otter species occur in sympatry (e.g., Southeast Asia; Koepfli et al., 2008).
One way to overcome these limitations is to employ molecular methods that provide powerful sources of species-specific as well as population- and individual-level information from various types of biological samples. In this context, the recent availability of whole-genome sequences from 13 of the 14 species in Lutrinae (de Ferran et al., 2022) has opened the possibility of performing surveys of microsatellite loci across this clade. In this study, we have employed this multi-taxon approach to identify novel microsatellite loci for 11 of the 14 otter species. We validated their efficiency by testing them on two species, providing a valuable resource that should help enhance genetic and ecological studies targeting these organisms.
MATERIAL AND METHODS
Genomic Dataset
To conduct a large-scale, standardized survey of microsatellite loci in the subfamily Lutrinae, we analyzed whole-genome sequences from 13 extant otter species. For 11 species (Aonyx cinereus, Aonyx capensis, Aonyx congicus, Hydrictis maculicollis, Lontra canadensis, Lontra felina, Lontra longicaudis, Lontra provocax, Lutra lutra, Lutra sumatrana and Lutrogale perspicillata), we used genomes that were previously reported by our group (de Ferran et al., 2022), while for Pteronura brasiliensis and Enhydra lutris, we used publicly available genome assemblies (Beichman et al., 2019). For two species (Lutra sumatrana and Aonyx congicus), the genome sequences were generated from museum specimens, and therefore had much lower depth of coverage and more missing data than the other species (see de Ferran et al., 2022). All 13 genomes were mapped against the Eurasian otter (Lutra lutra) reference genome (Mead et al., 2020) following the protocol reported by de Ferran et al. (2022). We generated consensus sequences of all genomes using ANGSD 0.921 (Korneliussen et al., 2014) with the parameters doFasta= 2, doCounts= 1, explode= 1, setMinDepth= 10 and minMapQ= 20.
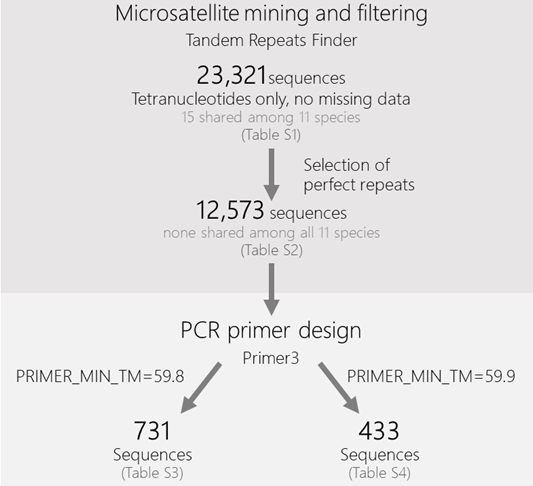
Microsatellite Mining and Filtering
To identify tandem repeats in each species’ genome, we used the Tandem Repeats Finder program (Benson, 1999) with default parameters: matching weight = 2, mismatching penalty = 7, indel penalty = 7, match probability = 80, indel probability = 10, minimum alignment score to report = 50, maximum period size to report = 4, to include flanking sequence (-f) and data file (-d), and maximum TR length expected in millions (-l) = 2. Considering the minimum score and the matching weight, the minimum number of required units in the tandem array was seven. We filtered the results stringently by keeping only tetranucleotides with complete and perfect repeats, and without any missing data in the target sequence or in either of the flanking regions (50 bases on each side of the repetitive array). Tetranucleotide loci were chosen due to their reduced stutter artifacts and simpler allele size determination (e.g., Edwards et al., 1991; Guichoux et al., 2011), despite potentially lower mutation rates compared to loci with smaller repeat motifs (e.g., dinucleotides; Chakraborty et al., 1997).
The repeat regions that were retained after these filtering steps were then assessed in terms of the efficiency of PCR primer design targeting their flanking sequences. Our aim was to develop microsatellite loci for each species with minimal need for PCR optimization by multiple end users (Robertson and Walsh-Weller, 1998). This was conducted with Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012) using the following parameters: PRIMER_TASK=generic, PRIMER_PICK_LEFT_PRIMER=1, PRIMER_PICK_INTERNAL_OLIGO=0, PRIMER_PICK_RIGHT_PRIMER=1, PRIMER_OPT_SIZE=20, PRIMER_MAX_SIZE=20, PRIMER_NUM_RETURN=1, PRIMER_OPT_TM=60.0, PRIMER_MAX_TM=60.1, PRIMER_MIN_TM=59.9, PRIMER_PRODUCT_SIZE_RANGE=80-400. To assess the effect of such a stringent range of melting temperatures (TM), we performed a second round of analysis with the same parameters, changing only PRIMER_MIN_TM to 59.8. We chose these temperature thresholds because of the increased specificity of PCR during primer annealing at higher temperatures (Roux, 1995; Hecker and Roux, 1996).
In addition to discovering microsatellite repeats in each species, we also assessed whether the identified loci were shared among different otter species. To do this, we compared the genomic coordinates of the identified repeats, since all species had their genome sequence consensus constructed using the same Eurasian otter reference genome assembly. We also compared the repeat motif and the flanking sequences of each shared locus to assess their variation across species.
Empirical Validation of Microsatellite Genotyping
To ascertain that microsatellites identified with our approach could be reliably genotyped, we empirically tested the markers developed for two Asian species, the smooth-coated otter (Lutrogale perspicillata) and the small-clawed otter (Aonyx cinereus) for which samples had been collected and were available to be used for this study. We optimized the 59.9 °C set of primers designed for each of these species using genomic DNA from only one individual of each, isolated previously with Qiagen DNeasy Blood & Tissue DNA isolation kits (www.qiagen.com) from tissue samples obtained from the cryo-collection of the Lee Kong Chian Natural History Museum, Singapore (lkcnhm.nus.edu.sg; Lutrogale perspicillata sample LKCNHM 119390 and Aonyx cinereus sample LKCNHM 118524). Additionally, we tested a set of 27 primers (7 from Aonyx cinereus, 11 from Lutrogale perspicillata, and 9 duplicated, occurring in both species) designed based on a random set of imperfect repeats, to assess whether they can also yield useful markers (Table S5).
All PCRs were conducted in 20 µL reaction volumes, using 10 µL Promega 2x Taq master mix (www.promega.com), 2 µL DNA eluate, and 1.0 µl each of 10 µM forward and reverse primers, on a Bio-Rad T100 thermocycler (www.bio-rad.com). First, we conducted a touchdown PCR with the following thermal cycling profile: 94 ºC for 180s as an initial denaturation step, followed by 30 cycles of 30s at 94 ºC for denaturation, 30s of annealing temperatures (Ta) starting at 65 °C and dropping 0.5 °C per cycle, and 30s at 72 ºC for extension, followed by 20 similar cycles with Ta of 50 ºC, then 240s at 72 ºC, and 4 ºC forever. PCR products were run on a 1% agarose gel to assess quality and were classified as either no product, multiple banding pattern indicative of non-specific binding, or a clear single-band PCR product (N, M, or Y, respectively). If we obtained a clear PCR product, or if a multiple banding pattern was not severe, we conducted gradient PCR to determine the optimum annealing temperature, with 30 cycles of 30s each of 94 ºC, Ta, and 72 ºC, where the gradient annealing temperatures ranged from 65 ° to 50 °C. We conducted the final PCRs using similar thermal cycling conditions to the gradient reaction conditions, at the optimized annealing temperature for each primer pair, and using forward primers labelled with fluorophores 6-FAM, HEX or TET. These PCR products were submitted to 1st-Base Asia, Singapore (.base-asia.com) where they were run on an Applied Biosystems capillary sequencer (https://www.thermofisher.com). Microsatellite allele sizes were scored using the Microsatellite v1.4.7 plugin in the Geneious Prime software (https://www.geneious.com).
RESULTS AND DISCUSSION
Using enrichment, cloning, and Sanger sequencing of genomic libraries, limited numbers of microsatellite loci have been previously characterized in several otter species including Lutra lutra (Dallas and Piertney, 1998; Huang et al., 2005), Enhydra lutris (Kretschmer et al., 2009), Lontra canadensis (Beheler et al., 2005) and Pteronura brasiliensis (Ribas et al., 2011). Microsatellite loci are abundant in eukaryotic genomes (Chantzi and Georgakopoulos-Soares, 2024). These loci can be efficiently identified from genome assemblies or sequencing data using bioinformatics tools like Tandem Repeats Finder (Benson, 1999). In this study, we identified a total of 23,321 tetranucleotide microsatellite loci within the genomes of 12 of the 13 otter species we analyzed (all but A. congicus). This total was obtained by summing the identified loci in each species, and therefore it includes markers that were duplicated due to being found in more than one species (Table S1). Among these loci, due to the enforcement of stringent filtering criteria, only 15 were shared among the 11 species with higher depth of coverage (i.e., excluding the genomes generated from museum samples, Table 1, Table S2). Of the two otters represented by museum samples, only one locus was identified for L. sumatrana, and none for A. congicus, indicating that the search criteria that we enforced were too stringent for lower-coverage genomes with larger amounts of missing data. However, it is noteworthy that each of them has a sister-species (L. lutra and A. capensis, respectively) that yielded a large amount of identified loci, which are likely to be applicable on samples obtained from L. sumatrana and A. congicus.
| Table 1: Total number of identified microsatellite loci per species for the 11 species with higher-coverage genomes, and the subsets for which PCR primers could be successfully designed with two different settings for the minimum melting temperature, Tm (expressed in °C). The single locus identified for L. sumatrana (see text) is not shown (see Tables S1, S2). | |||||
| Species | Total | Only Perfect Repeats | Successful Primer Design. Minimum Tm=59.8 °C |
Successful Primer Design. Minimum Tm=59.9 °C |
|
| Aonyx cinereus | 2,157 | 1,136 | 56 | 28 | |
| Aonyx capensis | 2,232 | 1,133 | 67 | 42 | |
| Enhydra lutris | 2,436 | 1,332 | 85 | 47 | |
| Hydrictis maculicollis | 1,590 | 794 | 44 | 27 | |
| Lontra canadensis | 1,556 | 854 | 39 | 28 | |
| Lontra felina | 1,182 | 610 | 32 | 16 | |
| Lontra longicaudis | 2,079 | 1,180 | 83 | 54 | |
| Lontra provocax | 1,027 | 510 | 23 | 12 | |
| Lutra lutra | 5,359 | 3,080 | 179 | 105 | |
| Lutrogale perspicillata | 2,308 | 1,213 | 66 | 43 | |
| Pteronura brasiliensis | 1,394 | 731 | 57 | 31 | |
| Total | 23,320 | 12,573 | 731 | 433 | |
Because some of the loci presented occasional differences in the repeat motif, which could be exclusive to one species or occur in multiple species (Fig. 1), we applied another filter layer to exclude these cases, leaving only perfect repeat arrays to be considered in the downstream step of primer design. Still, because these variations may be of interest to researchers working on these species, these data are also included in the Supplementary Material. After this new filter, there were 12,573 sequences left (Table 1; Fig. 2; Table S2), with no locus left for L. sumatrana, and no shared loci among the 11 remaining species (Table S2). At the same time, if a smaller set of species was assessed (e.g., those occurring on the same continent), some overlap in the retrieved loci could be discerned, opening the possibility of designing markers that can be applied to sets of sympatric otter species.

The primer design step was completed successfully for 731 sequences (representing 420 unique loci) for the 59.8 °C minimum melting temperature (Table S3) and 433 sequences (representing 259 unique loci) for the 59.9 °C minimum melting temperature (Table 1, Table S4). This substantial difference in the number of retrieved loci with only a change of 0.1 °C in the minimum Tm illustrates the impact of this parameter when performing such genome-wide surveys of microsatellite markers and highlights the need to consider it carefully. Overall, the 59.9 °C minimum melting temperature group presented 57 repeat patterns, and the 59.8 °C one presented 68 patterns. For both, the most common repeat patterns were the ones containing AT combinations (AAAT, ATTT, TAAA, and TTTA).
Although we used a different Lutra lutra individual relative to the one employed for the reference genome (to minimize the potential influence of the mapping reference), Lutra lutra was the species with the largest number of identified loci. The reference seemed to influence the number of retrieved loci per species, with species closer to the reference (i.e., with a more recent divergence) having, overall, a larger number of identified loci and a lower amount of missing data (hence, fewer sequences were excluded during the filtering steps). This in turn led to the observed bias in the success rate for the reference species (see Table 1). In spite of this bias, we still obtained a substantial number of microsatellite loci for every surveyed species (>20 loci with primers designed with a minimum Tm=59.9 °C; >40 loci with a minimum Tm=59.8 °C), demonstrating the potential of this approach to identify such sequences across a whole group of related species, while enforcing strict criteria that maximize the species-specificity and informative potential of these markers.
We assessed the number of shared loci among species from the same continent, considering only markers for which PCR primer design was successful (Fig. 3). The observed trends were similar for both assessed melting temperatures, with several combinations of shared markers among potentially sympatric species. For example, in Eurasia, there were 13 shared loci among the three surveyed species when the minimum Tm was 59.9 °C, and 20 shared loci with minimum Tm=59.8 °C. In South America, where four species were compared, different combinations of shared loci could be retrieved. Among these, the two sympatric tropical species Pteronura brasiliensis and Lontra longicaudis shared 10 loci with minimum Tm=59.8 °C, exemplifying a marker set that can be useful to investigate both species in the field.
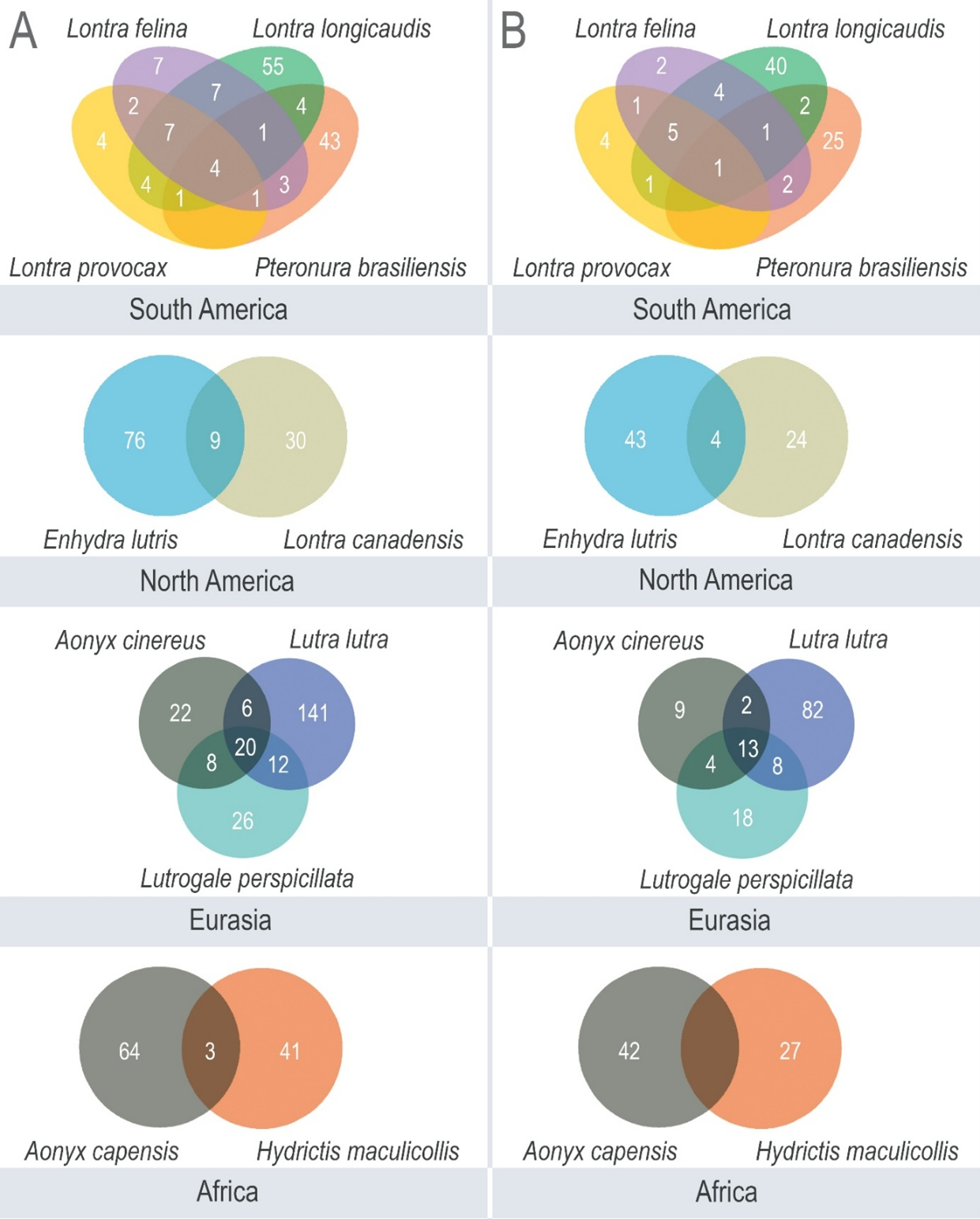
Finally, the marker set with minimum Tm of 59.9 °C identified for Aonyx cinereus (44 in total) and Lutrogale perspicillata (63 in total), based on perfect and imperfect repeats (Table S4, S5), was empirically tested in one individual of each of these species. Considering that these species had primers in common (Fig. 3), the total number of loci/primer sets tested was 81 (18 designed for A. cinereus, 37 designed for L. perspicillata, and 26 common to both). Of these, 51 yielded successful microsatellite genotypes in both species (Table S6), and in 34 of them we observed heterozygosity in at least one of the otters (Table S6). Interestingly, of the nine loci designed exclusively for A. cinereus that showed variability, two were heterozygous in the target species and homozygous in L. perspicillata, while four showed the opposite pattern and three were heterozygous in both species. Similarly, of the 12 loci designed exclusively for L. perspicillata that showed variability, four were heterozygous in the target species and homozygous in A. cinereus, while five showed the opposite pattern and three were heterozygous in both species. We did not find any evident difference regarding the allelic size range between heterologous and homologous loci. These results demonstrate that these markers can be successfully applied in empirical genotyping, and that they are informative (i.e., variable) not only in their respective target species but also in another, closely related otter. However, we acknowledge that comparing monomorphic or polymorphic loci across multiple individuals of the two species would be more informative than comparing a single homozygous or heterozygous individual between these species.
Similar sets can be constructed for other combinations of otter species that occur in a given area, not only using the Tm range that we employed here, but also considering a broader suite of PCR parameters, starting from our complete list of identified tetranucleotide markers (Tables S1, S2). In addition, microsatellite marker sets can be designed to allow for multiplexing, thereby making data collection more efficient (Puckett, 2017). Furthermore, when paired with markers containing species-specific diagnostic sites (e.g., Koepfli et al., 2008), our panel of microsatellite loci will empower genetic and population monitoring of otter species with overlapping distributions. The microsatellite loci we report in this study will need to be empirically assessed for polymorphism and information content by individual researchers and if they are applied to non-invasive samples such as hair and spraints, further optimization and testing may be required. We believe this resource should be useful to facilitate global research on otter ecology and genetics, accelerating the collection of field data with relevant implications for conservation planning on behalf of these species.
Acknowledgements: We thank Foo Maosheng (Lee Kong Chian Natural History Museum, Singapore) for access to samples. Financial support for this study was provided by CNPq/Brazil (grants 141172/2017-7 [awarded to V.d.F.] and 309068/2019-3 [awarded to E.E.]), the PUCRS/CAPES-PrInt Program (fellowship 88887.370464/2019-00 awarded to V.d.F.), the Ministry of Education through the Yale-NUS College start-up grant R-607-265-226-121 (awarded to C.E.E.C. and P.J.) and by the Yale-NUS College Centre for International & Professional Experience (awarded to C.E.E.C.). This study is a contribution of the National Institutes for Science and Technology (INCT) in Ecology, Evolution and Biodiversity Conservation, supported by MCTIC/CNPq/Brazil (proc. 465610/2014-5) and FAPEG/Brazil (proc. 201810267000023). We thank two anonymous reviewers who provided constructive comments on an earlier version of this manuscript.
REFERENCES
Beheler, A.S., Fike, J.A., Dharmarajan, G., Rhodes Jr, O.E., Serfass, T.L. (2005). Ten new polymorphic microsatellite loci for North American river otters (Lontra canadensis) and their utility in related mustelids. Mol. Ecol. Notes 5(3): 602-604. https://doi.org/10.1111/j.1471-8286.2005.01005.x
Beichman, A.C., Kalhori, P., Kyriazis, C.C., DeVries, A.A., Nigenda-Morales, S., Heckel, G., Schramm, Y., Moreno-Estrada, A., Kennett, D.J., Hylkema, M., Bodkin, J., Koepfli, K-P., Lohmueller, K.E., Wayne, R.K. (2023). Genomic analyses reveal range-wide devastation of sea otter populations. Mol. Ecol. 32(2): 281–298. https://doi.org/10.1111/mec.16334
Beichman, A.C., Koepfli, K-P., Li, G., Murphy, W., Dobrynin, P., Kilver, S., Tinker, M.T., Murray, M.J., Johnson, J., Lindblad-Toh, K., Karlsson, E.K., Lohmueller, K.E., Wayne, R.K. (2019). Aquatic adaptation and depleted diversity: A deep dive into the genomes of the sea otter and giant otter. Mol. Biol. Evol. 36(12): 2631–2655. https://doi.org/10.1093/molbev/msz101
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2): 573–580. https://doi.org/10.1093/nar/27.2.573
Bhat, N.N., Mahiya-Farooq, Padder, B.A., Shah, M.D., Dar, M.S., Nabi, A., Bano, A., Rasool, R.S., Sana-Surma (2018). Microsatellite mining in the genus Colletotrichum. Gene Rep. 13: 84–93. https://doi.org/10.1016/j.genrep.2018.09.001
Chakraborty, R., Kimmel, M., Stivers, D.N., Davison, L.J., Deka, R. (1997). Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl. Acad. Sci. USA 94: 1041-1046. https://doi.org/10.1073/pnas.94.3.1041
Chantzi, N., Georgakopoulos-Soares, I. (2024). The repertoire of short tandem repeats across the tree of life. bioRxiv 2024.08.08.607201. https://doi.org/10.1101/2024.08.08.607201
Crowley, S., Johnson, C.J., Hodder, D. (2012). Spatial and behavioral scales of habitat selection and activity by river otters at latrine sites. J. Mamm. 93(1): 170–182. https://doi.org/10.1644/10-MAMM-A-362.1
Dallas, J.F., Piertney, S.B. (1998). Microsatellite primers for the Eurasian otter. Mol. Ecol. 7(9): 1248-1251. https://chemport-n.cas.org//chemport-n/?APP=ftslink&action=reflink&origin=npg&version=1.0&coi=1%3ACAS%3A528%3ADyaK1cXmtFOisbc%3D&md5=dc0b47bbe9d547e56aae6156820f549e
de Ferran, V., Figueiró, H.V., Trinca, C.S., Hernández-Romero, P.C., Lorenzana, G.P., Gutiérrez-Rodríguez, C., Koepfli, K-P., Eizirik, E. (2024). Genome-wide data support recognition of an additional species of Neotropical river otter (Mammalia, Mustelidae, Lutrinae). J. Mamm. 105(3): 534–542. https://doi.org/10.1093/jmammal/gyae009
de Ferran, V., Figueiró, H.V., Trindade, F.J., Oliver Smith, Sinding, M.H.S., Trinca, C.S., Lazzari, G.Z., Veron, G., Vianna, J.A., Barbanera, F., Kliver, S., Serdyukova, N., Bulyonkova, T., Ryder, O.A., Gilbert, M.T.P., Koepfli, K-P., Eizirik, E. (2022). Phylogenomics of the world’s otters. Curr. Biol. 32(16): 3650-3658. https://doi.org/10.1016/j.cub.2022.06.036
Edwards, A., Civitello, A., Hammond, H.A., Caskey, C.T. (1991). DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet. 49(4), 746–756. https://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC1683171&blobtype=pdf
Frankham, R., Ballou, J.D., Briscoe, D.A. (2002). Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK https://doi.org/10.1017/CBO9780511808999
Fuentes-Pardo, A.P., Ruzzante, D.E. (2017). Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 26(20): 5369-5406. https://doi.org/10.1111/mec.14264
Guichoux, E., Lagache, L., Wagner, S., Chaumeil, P., Léger, P., Lepais, O., Lepoittevin, C., Malausa, T., Revardel, E., Salin, F., Petit, R.J. (2011). Current trends in microsatellite genotyping. Mol. Ecol. Resour. 11(4): 591-611. https://doi.org/10.1111/j.1755-0998.2011.03014.x
Harper, C., Ludwig, A., Clarke, A., Kakgopela, K., Yurchenko, A., Guthrie, A., Dobrynin, P., Tamazian, G., Emslie, R., van Heerden, M., Hofmeyr, M., Potter, R., Roets, J., Beytell, P., Otiende, M., Kariuki, L., du Toit, R., Anderson, N., Okori, J., Antonik, A., Koepfli, K-P., Thompson, P., O’Brien, S.J. (2018). Robust forensic matching of confiscated horns to individual poached African rhinoceros. Curr. Biol. 28(1): R13-R14. https://doi.org/10.1016/j.cub.2017.11.005
Hauser, S.S., Athrey, G., Leberg, P.L. (2021). Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Ecol. Evol. 11(22): 15800-15814. https://doi.org/10.1002/ece3.8250
Hecker, K.H., Roux, K.H. (1996). High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques 20(3): 478-85. https://doi.org/10.2144/19962003478 . PMID: 8679209
Huang, C-C., Hsu, Y-C, Lee, L-L., Li, S-H. (2005). Isolation and characterization of tetramicrosatellite DNA markers in the Eurasian otter (Lutra lutra). Mol. Ecol. Notes 5(2): 314-316. https://doi.org/10.1111/j.1471-8286.2005.00912.x
Ishida, Y., Gugala, N.A., Georgiadis, N.J., Roca, A.L. (2018). Evolutionary and demographic processes shaping geographic patterns of genetic diversity in a keystone species, the African forest elephant (Loxodonta cyclotis). Ecol. Evol. 8: 4919–4931. https://doi.org/10.1002/ece3.4062
IUCN (2022). The IUCN Red List of Threatened Species. Version 2021-3. Available at: https://www.iucnredlist.org
Koepfli, K-P., Kanchanasaka, B., Sasaki, H., Jacques, H., Louie, K.D.Y., Hoai, T., Dang, N.X., Geffen, E., Gutleb, A., Han, S-Y., Heggberget, T.M., LaFontaine, L., Lee, H., Melisch, R., Ruiz-Olmo, J., Santos-Reis, M., Sidorovich, V.E., Stubbe, M., Wayne, R.K. (2008). Establishing the foundation for an applied molecular taxonomy of otters in Southeast Asia. Conserv. Genet. 9: 1589-1604. https://doi.org/10.1007/s10592-007-9498-5
Koressaar, T., Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10): 1289-91. https://doi.org/10.1093/bioinformatics/btm091
Korneliussen, T.S., Albrechtsen, A., Nielsen, R. (2014). ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15: 356. https://doi.org/10.1186/s12859-014-0356-4
Kretschmer, E.J., Olsen, J.B., Wenburg, J.K. (2009). Characterization of eight microsatellite loci in sea otter, Enhydra, lutris, and cross-species amplification in other Mustelidae. Conserv. Genet. 10(3): 775–777. https://doi.org/10.1007/s10592-008-9660-8
Kumari, R., Wankhede, D.P., Bajpai, A., Maurya, A., Prasad, K., Gautam, D., Rangan, P., Latha, M., John, K.J., Suma, A., Bhat, K.V., Gaikwad, A.B. (2019). Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): A valuable resource for boosting genomics application ns. PLoS ONE 14(12): e0226002. https://doi.org/10.1371/journal.pone.0226002
Larson, S., Gagne, R.B., Bodkin, J., Murra,y M.J., Ralls, K., Bowen L., Leblois, R., Piry, S., Penedo, M.C., Tinker, M.T., Ernest, H.B. (2021). Translocations maintain genetic diversity and increase connectivity in sea otters, Enhydra lutris. Mar. Mammal. Sci. 37: 1475–1497. https://doi.org/10.1111/mms.12841
Latorre-Cardenas, M.C., Gutiérrez-Rodríguez, C., Lance, S.L. (2020). Isolation and characterization of 13 microsatellite loci for the Neotropical otter, Lontra longicaudis, by next generation sequencing. Mol. Biol. Rep. 47: 731–736. https://doi.org/10.1007/s11033-019-05165-z
Li, B., Kimmel, M. (2013). Factors influencing ascertainment bias of microsatellite allele sizes: impact on estimates of mutation rates. Genetics 195(2): 563–572. https://doi.org/10.1534/genetics.113.154161
Liu, S., Hou, W., Sun, T., Xu, Y., Li, P., Yue, B., Fan, Z., Li, J. (2017). Genome-wide mining and comparative analysis of microsatellites in three macaque species. Mol. Genet. Genom. 292: 537–550. https://doi.org/10.1007/s00438-017-1289-1
Liu, W., Xu, Y., Li, Z., Fan, J., Yang, Y. (2019). Genome-wide mining of microsatellites in king cobra (Ophiophagus hannah) and cross-species development of tetranucleotide SSR markers in Chinese cobra (Naja atra). Mol. Biol. Rep. 46: 6087–6098. https://doi.org/10.1007/s11033-019-05044-7
Manee, M.M., Algarni, A.T., Alharbi, S.N., Al-Shomrani, B.M., Ibrahim, M.A., Binghadir, S.A., Al-Fageeh, M.B. (2020). Genome-wide characterization and analysis of microsatellite sequences in camelid species. Mamm. Res. 65: 359–373. https://doi.org/10.1007/s13364-019-00458-x
Mead, D., Hailer, F., Chadwick, E., Portela Miguez, R., Smith, M., Corton, C., Oliver, K., Skelton, J., Betteridge, E., Doulcan, J.D., Dudchenko, O., Omer, A., Weisz, D., Lieberman Aiden, E., McCarthy, S., Howe, K., Sims, Y., Torrance, J., Tracey, A., Challis, R., Durbin, R., Blaxter, M. (2020). The genome sequence of the Eurasian river otter, Lutra lutra Linnaeus 1758. Wellcome Open Res. 5: 33. https://doi.org/10.12688/wellcomeopenres.15722.1
Miller, S.M., Harper, C.K., Bloomer, P., Hofmeyr, J., Funston, P.J. (2014). Evaluation of microsatellite markers for populations studies and forensic identification of African lions (Panthera leo). J. Hered. 105(6): 856–866. https://doi.org/10.1093/jhered/esu054
Puckett, E.E. (2017). Variability in total project and per sample genotyping costs under varying study designs including with microsatellites or SNPs to answer conservation genetic questions. Conserv. Genet. Resour. 9: 289–304. https://doi.org/10.1007/s12686-016-0643-7
Ribas, C., Vasconcellos, A.V., Mourão, G., Magnusson, W., Solé-Cava, A.M., Cunha, H.A. (2011). Polymorphic microsatellite loci from the endangered Giant Otter (Pteronura brasiliensis). Conserv. Genet. Resour. 3(4): 769–771. https://doi.org/10.1007/s12686-011-9454-z
Rivera, N.A., Totoni, S., Monick, K., Tian, T., Green, M.L., Novakofski, J., Mateus-Pinilla, N.E. (2019). A comparison of three methods to evaluate otter latrine activity. Wildl. Soc. Bull. 43(1): 198-207. https://doi.org/10.1002/wsb.947
Robertson, J.M., Walsh-Weller, J. (1998). An introduction to PCR primer design and optimization of amplification reactions. Methods Mol. Biol. 98: 121-154. https://doi.org/10.1385/0-89603-443-7:121
Roux, K.H. (1995). Optimization and troubleshooting in PCR. PCR Methods Appl. 4: S185-S194. https://doi.org/10.1101/gr.4.5.s185
Schwartz, M.K., Luikart, G., Waples, RS. (2007). Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22(1): 25-33. https://doi.org/10.1016/j.tree.2006.08.009
Sun, N.C-M., Chang, S-P., Lin, J-S., Tseng, Y-W., Pei, K.J-C., Hung, K-H. (2020). The genetic structure and mating system of a recovered Chinese pangolin population (Manis pentadactyla Linnaeus, 1758) as inferred by microsatellite markers. Glob. Ecol. Conserv. 23: e01195. https://doi.org/10.1016/j.gecco.2020.e01195
Trinca, C.S., Jaeger, C.F., Eizirik, E. (2013). Molecular ecology of the Neotropical otter (Lontra longicaudis): non-invasive sampling yields insights into local population dynamics. Biol. J. Linn. Soc. 109(4): 932–948. https://doi.org/10.1111/bij.12077
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B.C., Remm, M., Rozen, S.G. (2012). Primer3--new capabilities and interfaces. Nucleic Acids Res. 40(15): e115. https://doi.org/10.1093/nar/gks596
Xu, Y., Li, W., Hu, Z., Zeng, T., Shen, Y., Liu, S., Zhang, X., Li, J., Yue, B. (2018). Genome-wide mining of perfect microsatellites and tetranucleotide orthologous microsatellites estimates in six primate species. Gene 643: 124-132. https://doi.org/10.1016/j.gene.2017.12.008
Excel Spreadsheet 3.6 MB https://iucnosgbull.org/Volume42/De_Ferran_et_al_2025_Supplementary_Information.xlsx
This contains six sheets as follows:
Table S1 - List of 23,321 microsatellite loci identified across 12 of the surveyed otter species, including species name and abbreviation, genomic position based on the Lutra lutra reference genome (chromosome, starting and ending position), repeat motif, number of copies, percentage of each base, sequence and flanks.
Table S2 - List of 12,573 microsatellite loci considering only sequences with perfect repeats, including species name and abbreviation, genomic position based on the Lutra lutra reference genome (chromosome, starting and ending position), repeat motif, number of copies, percentage of each base, sequence and flanks.
Table S3 - List of 731 loci which passed the primer design step, considering minimum melting temperatures of 59.8.
Table S4 - List of 433 loci which passed the primer design step, considering minimum melting temperatures of 59.9.
Table S5 - Set of 27 primers designed based on a random set of imperfect repeats tested on one individual of Aonyx cinereus and one individual of Lutrogale perspicillata.
Table S6 - Results of a set of 51 primers successfully tested on one individual of Aonyx cinereus and one individual of Lutrogale perspicillata. Cells with red boldface indicate the 34 loci/primers that resulted in heterozygous genotypes in one or the other or both species.
Résumé: Identification à Grande Échelle de Loci Microsatellites chez Plusieurs Espèces de Loutres (Mammalia, Carnivora, Lutrinae) à l’Aide de Données de Séquences de Génomes Complets
Le développement d’études moléculaires sur des espèces discrètes, rares et/ou mal connues est confronté à des défis en raison du manque de marqueurs appropriés. Les marqueurs microsatellites spécifiques à l’espèce minimisent les biais, offrent de meilleures performances et sont rentables, facilitant le développement d’études de la génétique des populations. L’utilisation de séquences de génomes entiers permet le développement de marqueurs microsatellites spécifiques à l’espèce et leur étude chez des espèces étroitement apparentées, permettant ainsi la découverte de marqueurs partagés qui peuvent faciliter les études comparatives. Les Lutrinés comportent actuellement 14 espèces de loutres. Malgré leur état de conservation préoccupant, en raison de caractéristiques inhérentes à ces espèces qui rendent leur étude difficile par les méthodes traditionnelles, beaucoup d’entre elles ne disposent pas de données génétiques de population fiables, ce qui limite les efforts de conservation. Dans cette étude, nous avons utilisé une approche multi taxon pour identifier un grand nombre de nouveaux loci microsatellites pour 11 des 14 espèces de loutres, en évaluant si les loci identifiés étaient partagés entre les différents taxons. Nous avons identifié un total de 23.320 loci microsatellites chez 11 espèces, nombre qui a été réduit à 12.573 loci après un filtrage rigoureux. La conception des amorces a été réalisée avec succès pour 420 et 259 loci uniques, en considérant deux températures de fusion minimales. Nous avons validé l’efficacité des marqueurs en testant les 81 loci conçus pour deux espèces asiatiques. Parmi ceux-ci, 51 ont produit des génotypes microsatellites fiables chez ces deux espèces, 34 présentant une variation allélique chez au moins l’une d’entre elles. Ces résultats démontrent que ces marqueurs sont applicables à un génotypage empirique à la fois pour leur cible et celles qui sont proches.
Revenez au dessus
Resumen:Identificación Aa Gran Escala de Loci de Microsatélites de Múltiples Especies de Nutria (Mammalia, Carnivora, Lutrinae) utilizando Datos de Secuencias de Genoma Completo
El desarrollo de estudios moleculares en especies elusivas, raras, y/o poco conocidas enfrenta desafíos debido a la falta de marcadores apropiados. Los marcadores microsatelitales específicos de especie minimizan el sesgo, ofrecen mejor performance, y son costo-efectivos, ayudando al desarrollo de estudios de genética poblacional. El uso de secuencias de genoma completo permite el desarrollo de marcadores microsatelitales específicos de especie y su búsqueda en especies estrechamente relacionadas, posibilitando el descubrimiento de marcadores compartidos que pueden facilitar estudios comparativos. Los Lutrinae incluyen 14 especies existentes de nutria. A pesar de su preocupante status de conservación, y debido a las características inherentes de estas especies que hacen difícil su estudio mediante métodos tradicionales, muchas de ellas carecen de datos confiables de genética poblacional, lo que limita los esfuerzos de conservación. En este estudio, empleamos un enfoque multi-taxon para identificar un gran número de loci microsatelitales novedosos para 11 de las 14 especies de nutria, evaluando si los loci identificados eran compartidos entre diferentes taxa. Identificamos un total de 23.320 loci microsatelitales en esas 11 especies, que fueron reducidos a 12.573 después de aplicar estrictos criterios de filtrado. Completamos exitosamente el diseño de primers para 420 y 259 loci singulares, considerando dos temperaturas mínimas de fusión. Validamos la eficiencia de los marcadores testeando los 81 loci diseñados para dos especies Asiáticas. De éstos, 51 loci rindieron genotipos microsatelitales confiables en ambas especies, con 34 mostrando variación alélica en al menos uno de ellos. Estos resultados demuestras que estos marcadores son aplicables en la genotipificación empírica, tanto para la especie-objetivo como las estrechamente relacionadas.
Vuelva a la tapa