IUCN/SSC Otter Specialist Group Bulletin

©IUCN/SCC Otter Specialist Group
Citation: Teoh, W.Y., Woo, C.Y., Sharma, S., Ratnayeke, S., Yow, Y.-Y., Chew, J., and Abdul-Patah. P. (2024). Sex Determination of the Asian Small-Clawed Otter (Aonyx cinereus) and Smooth-Coated Otter (Lutrogale perspicillata) by DNA Analysis of Spraints. IUCN Otter Spec. Group Bull. 41 (2): 97 - 114
Sex Determination of the Asian Small-Clawed Otter (Aonyx cinereus) and Smooth-Coated Otter (Lutrogale perspicillata) by DNA Analysis of Spraints
Wan Yi Teoh1*, Chee Yoong Woo1,2, Sandeep Sharma3, Shyamala Ratnayeke1,5, Yoon-Yen Yow1, Jactty Chew1, and Pazil Abdul-Patah4
1Department of Biological Sciences, School of Medical and Life Sciences, Sunway University, 5, Jalan Universiti, Bandar Sunway, 47500 Petaling Jaya, Selangor, Malaysia
2Malaysian Nature Society, JKR 641, Jalan Kelantan, Bukit Persekutuan, 50480, Kuala Lumpur, Malaysia
3German Centre for Integrative Biodiversity Research (iDiv), Halle-Jena-Leipzig, Germany and Institute of Biology, Martin Luther University Halle-Wittenberg, Halle, Germany
3Department of Wildlife and National Parks (PERHILITAN), Peninsular Malaysia, Kuala Lumpur, Malaysia
3Allegany College of Maryland, Cumberland, MD 21502, USA
*Corresponding Author Email: teohwanyiii@gmail.com
Received 29th June 2023, accepted 6th January 2024
Abstract: Sex identification in natural populations provides insights into population demographics, species kin relationships, and behavioral strategies. We evaluated the applicability of two established sex markers, namely the sex determining region (SRY) gene and zinc finger (ZFX/ZFY) gene on the Asian small-clawed otter (Aonyx cinereus) and smooth-coated otter (Lutrogale perspicillata) in Malaysia. We used these primers to amplify a portion of the SRY and ZFX/ZFY genes to amplify the DNA extracted from tissue samples of otters of known sex and later tested their efficacy to amplify non-invasive samples with 20 spraint samples from wild otters. The amplicons were then observed by resolving on agarose gel. Results of DNA amplification of all six tissue samples accurately sex-typed otters for both markers. The SRY marker yielded a 70 bp product that amplifies only from males, whereas the ZFX/ZFY marker produced a single fragment of 180 bp in both sexes. Wild-collected spraint samples yielded amplicons of correct size for the SRY marker; however amplification success was low with the ZFX/ZFY marker where five of 20 samples produced the expected PCR product in at least two out of three replicates, five produced spurious bands, and no PCR products were detected with the remainder. Stringent laboratory techniques are required when dealing with non-invasive samples such as spraints where allelic drop-out and PCR products from non-target DNA are common problems. To further test the robustness of both markers on non-invasive samples, fecal samples of known sex collected from captive otters are recommended for future studies. In conclusion, both SRY and ZFX/ZFY sex markers performed reliably for Asian small-clawed and smooth-coated otters. The success of both sex markers suggests that this method is applicable in wildlife forensics and demographic studies of otters in Malaysia and elsewhere in their range.
Keywords: Sex-determining region gene SRY, Zinc finger protein gene ZFX/ZFY, Molecular sexing, Non-invasive sampling
INTRODUCTION
Amongst the 13 species of otters worldwide, three species, namely the Asian small-clawed otter (Aonyx cinereus), smooth-coated otter (Lutrogale perspicillata), and hairy-nosed otter (Lutra sumatrana), are reported in various freshwater and coastal ecosystems in Peninsular Malaysia (Shariff, 1984; Burhanuddin and Ahmad, 1990; Hussain et al., 2011). Within Malaysia, the Asian small-clawed otter and smooth-coated otter are relatively common and classified as Least Concern, whereas the rare hairy-nosed otter is classified as Endangered (Sivasothi and Burhanuddin, 1994; Wildlife Conxervation Act, 2010). Although no reliable estimates of population size are available for these three species, global population trends are considered to be declining due to anthropogenic pressure (Khoo et al., 2021). Hence, both Asian small-clawed and smooth-coated otters are designated as globally Vulnerable by the International Union for Conservation of Nature (IUCN; Khoo et al., 2021; Wright et al., 2021).
Otters are emblematic species for wetland conservation. As apex predators in freshwater ecosystems, otters are sparsely dispersed in riverine landscapes, influencing trophic dynamics in this extensive system, including the vast flood plains of major rivers (Khan et al., 2014). The loss of otters from aquatic systems can affect food chains and ecosystem function (Estes and Palmisano, 1974). The deposition of otter spraints along shorelines facilitates nutrient transfer from freshwater to terrestrial systems (Ben-David et al., 2005). Because otters rely on a healthy prey base in aquatic ecosystems, their presence often serves as a biological indicator of wetland quality (Khan et al., 2014).
In Malaysia, very few studies have focused specifically to understand otter populations and their ecology, with most reports coming from bycatch data of other studies (e.g., roadkills, interviews with local aquaculturists) and ad hoc natural history observations (Tan, 2015; Salahshour, 2016; Ishigami et al., 2017; Pain, 2020; Wilson and Namaskari, 2020). Deficiencies in baseline data on the distribution, ecology and population genetics of otters are critical for conservation efforts of otters in Malaysia (Abdul-Patah et al., 2014). Research on population size and demographic characteristics, such as reproductive output and age and sex ratios, are fundamental to designing adaptive management strategies for threatened species (Nichols and Armstrong, 2012; DeMay et al., 2017).
Sex determination in natural populations provides valuable information on species kin relationships, demographic parameters (sex ratio), dispersal patterns and effective population size (Smith et al., 2006). Deviations from a 1:1 sex ratio (Fisher, 1930) often underlie life history strategies for maximizing reproductive success in many species (Dos Remedios et al., 2010). Knowledge of sex and sex ratios of individuals in a population allows for better preparation in conservation management (Taberlet et al., 1997). Skewed adult sex ratios, such as in polygamous mating systems, can greatly reduce effective population size (Nunney, 1993). In small, isolated populations, the negative effects of skewed sex ratios are far greater, reducing mating opportunities and genetic diversity in subsequent generations and increasing extinction risk (Lande 1993).
In recent decades, polymerase chain reaction (PCR)-based methods that target genetic differences between male and female individuals have successfully sex-typed otters and other mustelids (Hrovatin and Kunej, 2018). In general, targeted genes for sexing amongst mammals are divided into two major groups: Y-chromosome specific regions and homologous sections on both X and Y chromosomes (Hrovatin and Kunej, 2018). The SRY gene is one of the most popular Y-chromosome markers, located on the distal region of the short arm of the Y chromosome in mammals, and thus, amplification only occurs with male samples (Fechner., 1996). The second group of markers allows co-amplification of homologous sections on both X and Y chromosomes. The zinc finger (ZFX/ZFY) and amelogenin (AMEL) regions are two such widely studied markers in mammals found in both X and Y chromosomes (Hrovatin and Kunej, 2018). Sexing individuals with this type of marker is a robust approach because amplification can be seen in both sexes with PCR-RFLP analysis using just one primer pair (Aasen and Medrano, 1990). In felids, the Y-chromosome gene of amelogenin (AMELY) has a 20 bp deletion as compared to the X-chromosome gene (AMELX) (Pilgrim et al. 2005), resulting in different-sized amplicons (male: 194, 214; female: 214) that can be discerned directly with PCR products. In mustelids and sea otters, the amelogenin gene has not proved to be effective because the X and Y regions are monomorphic (Hattori et al., 2003). The ZFX/ZFY marker was successfully tested on several species of mustelids, including mink, ermine, marten, badger, and otter (Statham et al., 2007). Not long after, Mucci and Randi (2007) developed a novel PCR-RFLP system for Eurasian otters using the ZFX/ZFY marker, but with a shorter amplified segment of 180 base pairs. Shorter amplicons are more effective for non-invasive genotyping of samples derived from spraints where degradation may affect both DNA quantity and quality (Sharma et al., 2022).
In some species such as sea otters that display clear sexual dimorphism in body size or skull morphology, the sex of the otters can be identified through direct observation (Hernández-Romero et al., 2015; Ryazanov and Maminov, 1996). However, most otters occur at low densities, and are elusive and cryptic in nature, making species and sex identification via field observations quite challenging (Murphy et al., 2021). Non-invasively-collected biological samples, such as hair or feces, provide a more feasible approach for sex identification in such situations (e.g., Taberlet et al., 1993; Statham et al., 2007; Sharma et al., 2022). To initiate such a study on otters in Malaysia, a suitable sex marker is warranted, especially one that can be readily amplified from samples that may experience rapid DNA degradation in warm, humid tropical conditions. No previous molecular sexing study has been performed on the Asian small-clawed otter or smooth coated otter. In this study, our aim was to evaluate the reliability of two sex-typing markers for both species with the use of established SRY and ZFX/ZFY primers.
MATERIALS AND METHODS
Sample collection and DNA extraction
The use of otter genetic specimens and experimental procedures were approved by the Sunway University Research Ethics Committee (Approval code: PGSUREC2020/ 063). Six genetic samples from tissues of road-killed otters (Table 1) were extracted using the GF-1® Tissue DNA Extraction Kits (Vivantis, Malaysia). We also extracted DNA from 20 scats/ anal jelly samples (n = 10, for each species) collected from different sites along the North-Central Selangor Coast (NCSC). These samples were previously extracted within four months of collection using the GF-1® Soil Sample DNA Extraction Kits (Vivantis, Malaysia) for an ongoing study on otters and the extracted DNA was processed using a PCR-RFLP protocol to ascertain the species (Sharma et al., 2022). We did not quantify DNA, but all samples used in this study were successfully amplified using a primer that targeted the mitochondrial D-loop region, namely TanaD-mod (Sharma et al., 2022), that yielded an estimated 200 bp product. Amplification products were subsequently digested with restriction enzymes to generate species-specific RFLP profiles.
| Table 1: Number of tissue and fecal samples used from both species. | ||||
| Species | Known sex (Tissue samples) | Unknown sex (Fecal samples) | ||
| Male | Female | |||
| L. perspicillata | 1 | 1 | 10 | |
| A. cinereus | 2 | 2 | 10 | |
PCR Amplification
In this study, we amplified two different sex-determining genes, namely the Y-chromosome specific SRY gene and Zinc Finger Region (ZFX/ZFY) gene. An estimated size of 70 bp of the SRY region was amplified with the primer set designed by Dallas et al. (2000) from L. lutra SRY gene sequence (Table 2). A total PCR mixture of 15 µL consisted of 3 µL of tissue DNA or 5.5 µL of spraint DNA, 7.5 µL of 2x ExPrime Taq Premix (GENETBIO Inc., Korea), 0.5 µM primers, and distilled water was added to the final volume. PCR was carried out using T100® Thermal Cycler (Bio‐Rad Laboratories) with the thermal cycle as following: 90 ℃/1 min 45 s, 20 cycles of [90 ℃/15 s, 60 ℃ – 0.5 ℃ per cycle/15 s], 15 cycles of [90 ℃/15 s, 50 ℃/15 s] and final extension of 72 ℃/1 min (Dallas et al., 2000).
| Table 2 : Sequences of forward (F) and reverse (R) oligonucleotide primers used in this study and the GenBank accession number of the DNA sequence used in primer design |
||||
| Gene | Primers | Sequence (5’ – 3’) | GeneBank Accession No. | |
| SRY 1 | Lut-SRY-F Lut-SRY-R |
GAATCCCCAAATGCAAAACTC GGCTTCTGTAAGCATTTTCCAC |
AB491588.1 | |
| ZFX/ZFY 2 | P1-5EZ ZFXYRb |
ATAATCACATGGAGAGCCACAAGCT TTGTTCAGCTGTCTCATATTCACA |
ZFX: EF409419.1 ZFY: F409420.1 |
|
| 1Developed by Dallas et al. (2000) 2Developed by Mucci and Randi (2007) |
||||
The second primer pair (Table 2) was used to amplify a conserved region of approximately 180 bp of the ZFX/ZFY gene (Mucci and Randi, 2007). PCR reaction volume of 25 µL contained 5 µL of tissue DNA or 9 µL of spraint DNA, 12.5 µL of 2x ExPrime Taq Premix (GENETBIO Inc., Korea), 0.75 µM primers, and distilled water added to reach the final volume. The PCR parameter was 94 ℃/2 min, 40 cycles of [94 ℃/30s, 58.2 ℃/30 s, 72 ℃/45 s] and a final extension of 72 ℃ for 10 min. Amplicons were resolved through gel electrophoresis on agarose gels stained with 1 uL of Atlas ClearSight DNA stain (BioAtlas, Estonia) and visualized under G: BOX Chemi XX9 gel box (Syngene, US). SRY amplicons were visualized on a 2% agarose gel, while ZFX/ZFY amplicons were visualized on a of 2.5% agarose gel. A slightly higher concentration of gel was used for ZFX/ZFY amplicons for better separation of DNA fragments.
RESULTS
Validation of SRY and ZFX/ZFY Sex Markers
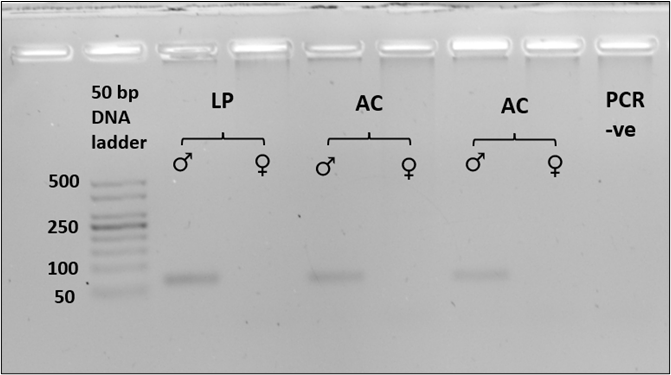
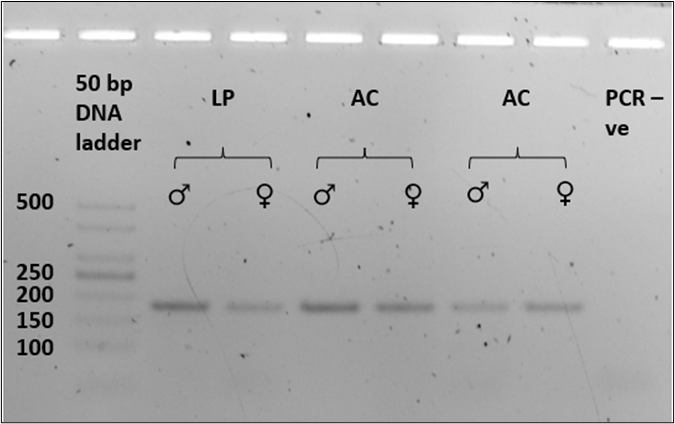
All six known-sex tissue samples were sex-typed based on band size on an agarose gel. Amplification of the SRY gene with the Lut-SRY-F/R primers yielded an approximately 70 bp product from known male tissue samples (n = 3) and no product was detected from the tested female samples (n = 3; Figure 1a). For the ZFX/ZFY marker, P1-5EZ and ZFXYRb primers successfully amplified a single product of approximately 180 bp of the zinc finger region from both known male and female tissue samples (n = 3, each; Figure 1b). The fragment length for both SRY and ZFX/ZFY gene was similar in both species and was consistent in all triplicates (100%). The absence of sex polymorphism in fragment size was evident in the amplified zinc finger region, which provided the expected amplicon product of similar size in both male and female samples (Figure 1b).
Sex Typing of 20 Fecal Samples
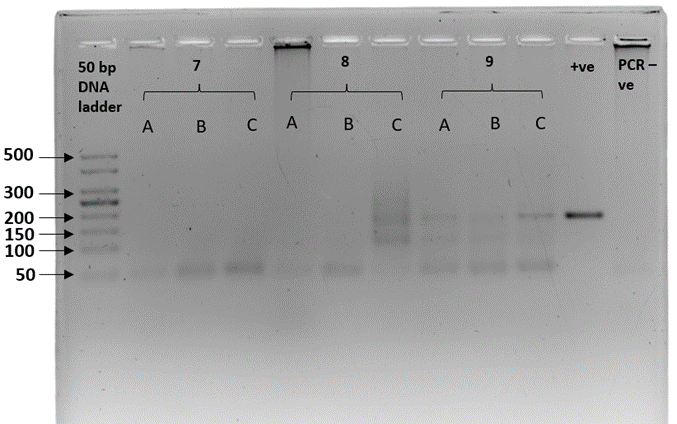
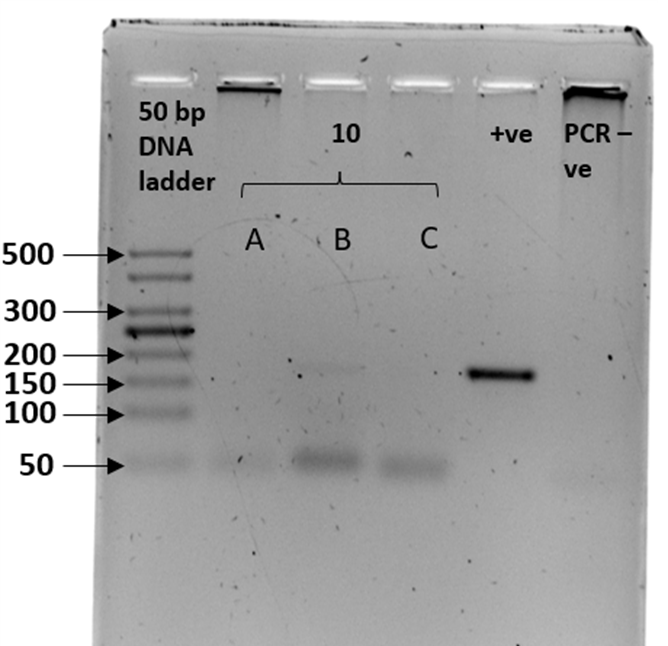
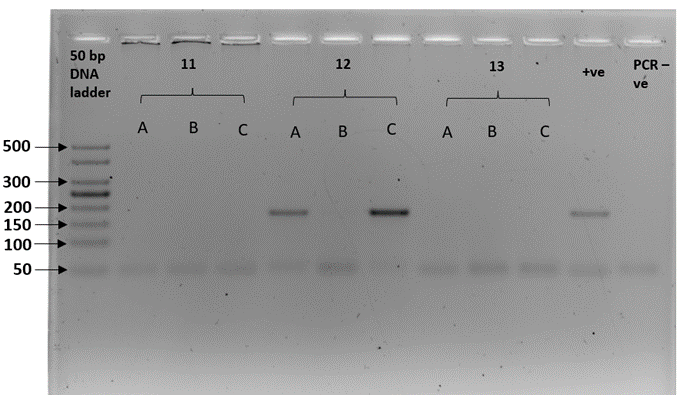
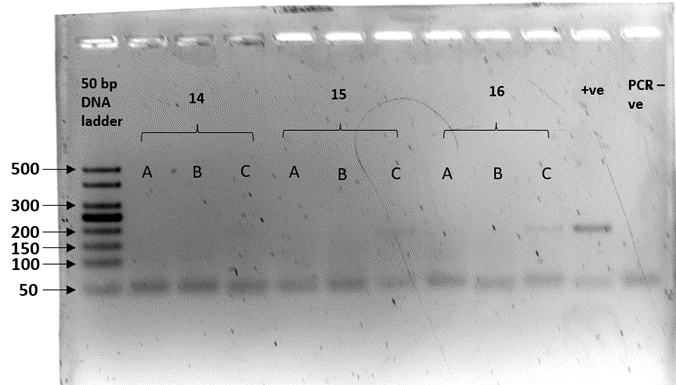
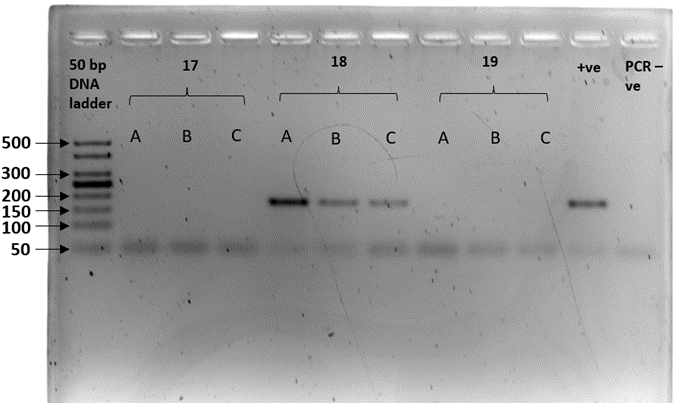
We sex-typed otter spraints collected from twenty different latrine sites with both SRY and ZFX/ZFY markers. Three PCR replicates were performed for all 20 fecal samples. The sex-typing result of a sample was accepted when there was a consistent result in at least two out of three PCR replicates. In SRY amplification, eight out of 20 fecal samples yielded a product of approximately 70 bp, concordant with the amplicon size recovered from the tissue samples (positive controls; Figure 2). Forty percent of the fecal samples were thus sex-typed as male. The 12 remaining samples (60%) with no amplified product of the expected size were tentatively identified as female (Table 3). Spurious amplification was seen in one smooth-coated otter fecal sample, KSJ 1-1 (Figure 2, Lane F3), with a product size of approximately 400 bp. Nevertheless, KSJ 1-1 was sex-typed as female due to the absence of expected SRY amplicon size for the otter. Overall, all samples exhibited consistent results across all three PCR replicates.
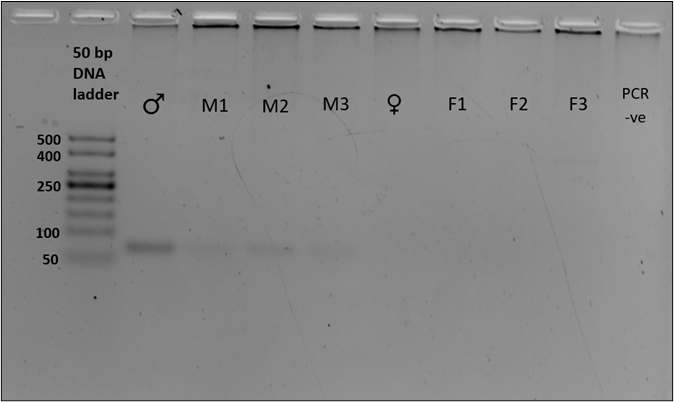
Amplification of the SRY region in fecal samples with the primers, Lut-SRY-F/R. M1, M2, and M3 were three out of the eight fecal samples that yielded an SRY amplicon at 70 bp, thus sex-typed as male. F1, F2, and F3 were identified as female due to the absence of expected product size. A faint band suggesting spurious amplification of approximately 400 bp was observed in F3. ♂ - male positive control; ♀ - female positive control; PCR -ve – non template control; M1 – SL 1-1; M2 – PB 4-1; M3 – SAT 1-(O)-1; F1 – BNO 6-1; F2 – PB 7-1; F3 – KSJ 1-1. Species identities for each sample ID are in Table 3.Click for larger version.
| Table 3: The sex determination results of 20 otter fecal samples used in this study were obtained by amplification of the sex-determining region (SRY) marker. Ten samples were from smooth-coated otters (LP) and ten from Asian small-clawed otters (AC). Male samples were identified by the presence of approximately 70 bp amplicons, whereas null amplification was sex-typed as female. LP – smooth-coated otter; AC – Asian small-clawed. Gel image is presented under Supplementary Figure 1. |
||||
| No. | Sample ID | Species | Sex | |
| 1 | SKB 3-1 | LP | M | |
| 2 | PB 4-1 | LP | M | |
| 3 | PB 9-1 | LP | F | |
| 4 | ST 36-1 | LP | F | |
| 5 | PB 10-2 | LP | M | |
| 6 | SN 1-1 | LP | F | |
| 7 | TAS 1-1 | LP | M | |
| 8 | SL 1-1 | LP | M | |
| 9 | KSJ 1-1* | LP | F | |
| 10 | PP 2-1 | LP | F | |
| 11 | KSNP 2-8 | AC | F | |
| 12 | SAT 1-(O)-2 | AC | M | |
| 13 | BNO 6-1 | AC | F | |
| 14 | PB 7-1 | AC | F | |
| 15 | PP 4-2 | AC | F | |
| 16 | PP 5-1 | AC | F | |
| 17 | PP 1-(O)-1B | AC | F | |
| 18 | PP 1-2(C) | AC | M | |
| 19 | PP 1-2 | AC | M | |
| 20 | KSL 3-1 | AC | F | |
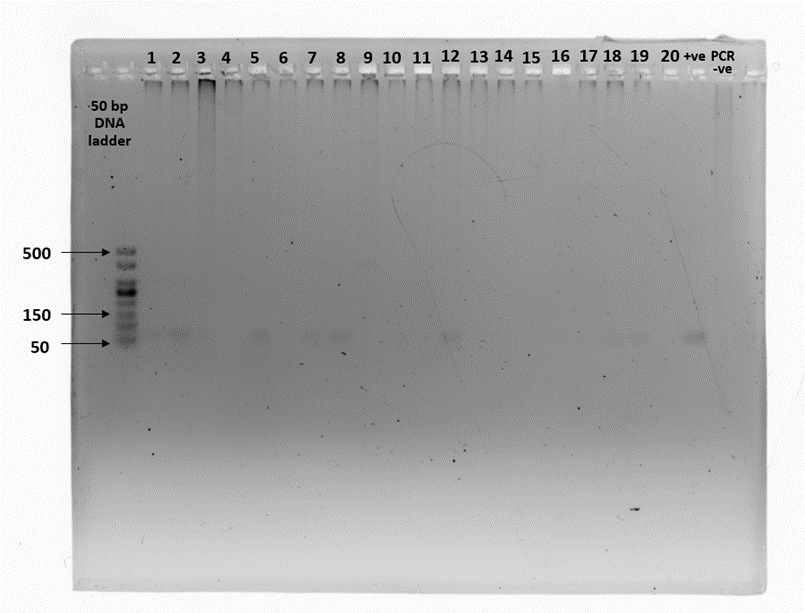
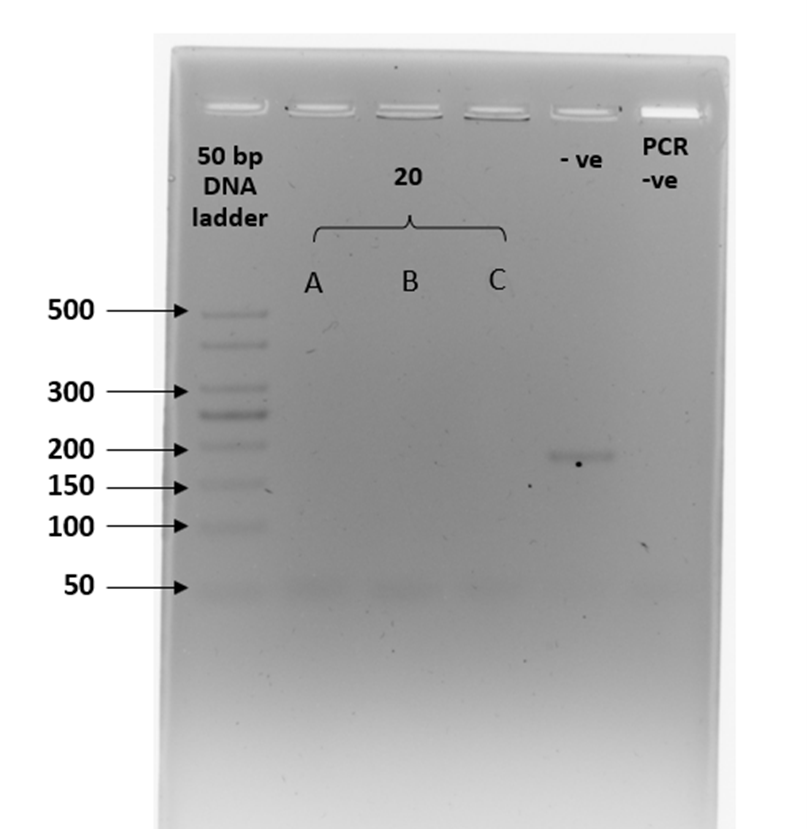
Amplification of the zinc finger region (ZFX/ZFY) was expected to produce a fragment of approximately 180 bp in all samples, regardless of sex. However, the expected amplicons were not evident in most of the fecal samples. Five out of 20 fecal samples (25%) displayed the expected amplification product in at least two out of three PCR replicates. Spurious bands occurred in five fecal samples from smooth-coated otters. Gel images of the three PCR replicates on 20 fecal samples are presented in Supplementary Figure 2 (a – h).
DISCUSSION
Validation of SRY and ZFX/ZFY Sex Markers
Both sex markers performed reliably during evaluation using tissue samples of known sex. For the SRY marker, amplification of an approximately 70 bp PCR product was displayed in all male tissue samples (n = 3; 1 from smooth-coated otter and 2 from Asian small-clawed otter) and were consistent across all three PCR replicates. SRY products of similar fragment length are also reported in several mustelids such as mink, ermine, marten, badger, Eurasian otter, and Neotropical otter (Statham et al., 2007; Trinca and Eizirik, 2012).
Smaller amplicon products such as the 70 bp product of the SRY gene, produce better results when used on degraded genetic samples (Statham et al., 2007). The major caveat with the SRY marker is that it only amplifies a male-specific region; therefore, null amplification of the SRY region could lead to inconclusive results (the sample could be a female otter, or the result of PCR failure). Amplification failures are common with non-invasive DNA samples (Fernando and Melnick, 2001), thus, the addition of an internal PCR positive control that amplifies a nuclear sequence is suggested to corroborate female samples (Forgacs et al., 2019). Unfortunately, an internal PCR control primer pair that could have served as a PCR positive control was unavailable in this study.
The amplification of ZFX/ZFY region is expected in all samples, regardless of sex. In this study, all tissue samples of known sex successfully yielded a single amplicon product of approximately 180 bp, matching the results obtained for the Eurasian otters (Lutra lutra) (Mucci and Randi, 2007; Park et al., 2011). A high degree of similarity in amplicon sizes of ZFX and ZFY is also reported in sex typing studies of Malayan tapir, horse, cattle, goats, sheep, and humans (Aaeson and Medrano, 1990; Palsboll et al., 1992; Lim et al., 2020). Hence, a suitable restriction enzyme is required after PCR to digest a suitable polymorphic site of the amplified ZFX/ZFY region so that the X and Y regions can be identified through different restriction fragment profiles (Hattori et al., 2003).
Shaw et al. (2003) demonstrated fragment size dimorphism between X (~900 bp) and Y (~1000 bp) using different pair of primers to target a larger portion of the zinc finger gene. However, large amplicons are likely to exhibit a higher failure rate when amplifying non-invasive samples (Statham et al., 2007). A maximum fragment size of 200 - 250 bp has been recommended, which improves the potential for restriction site polymorphisms in comparison with smaller amplicon products (Pages et al., 2009; Murakami et al., 2001).
Sex Typing of 20 Wild Fecal Samples
In this study, eight out of 20 samples were sex-typed as male using the 70 bp SRY marker, and all three replicates were consistent. For the 180 bp ZFX/ZFY marker, amplification success was 25% (five out of 20 fecal samples), where at least two out of three replicates gave consistent results. This success rate is comparatively lower than the 60% success rate reported in a study on Eurasian otters (Park et al., 2011). Panasci et al. (2011) found that amplification success and error rates were inversely proportional to sample age. Park et al. (2011) selectively tested fresh otter fecal samples, whereas the age of fecal samples used in our study was uncertain.
The approach we used in this study allows for sex-typing only with the SRY marker. The ZFX/ZFY marker produces a monomorphic product regardless of sex. Thus, the ZFX/ZFY marker could potentially serve as an internal positive control along with the SRY marker in a duplex PCR reaction. Thus, females can be identified with certainty in cases where ZFX/ZFY yields the expected product (180 bp) along with null amplification of the SRY product (70 bp). The presence of both products confirms that the sample is male. The incorporation of two sex markers, employing the zinc finger region as an internal control, is reported in sex-typing of the Malayan tapir (Lim et al., 2020).
The spurious amplification of an approximately 400 bp fragment was observed in one of the fecal samples using SRY markers (Figure 2, Lane F3), including five fecal samples using the ZFX/ZFY marker, although this did not impede sex-typing, because band sizes differed markedly from the otter-specific fragments. Mucci and Randi (2007) report the amplification of DNA of common prey species of the Eurasian otter with the ZFX/ZFY markers. Band sizes were well outside the range of those produced by Eurasian otters, except for one band (176 bp) produced by Cyprinidae fish. Murphy et al. (2003) pointed out that fecal samples of many species of carnivores are prone to similar kinds of amplification errors because of their diet. Otters are carnivorous, with a diet comprising mostly fish and crustaceans (Theng et al., 2016; Hon et al., 2010). Thus, the probability that sex markers may also amplify prey DNA cannot be discounted. In this study, spurious amplification only arose with samples from smooth-coated otters that are specialized fish-eaters, whereas Asian small-clawed otters feed mainly on crustaceans such as crabs (Kruuk et al., 1994).
Fecal samples are one of the most easily available, non-invasive sources for collecting genetic data from the wild (Zhu et al., 2017). Because of their elusive nature, monitoring programs on otters rely heavily on the use of spraint (Yoxon and Yoxon, 2014), but fecal amplification success is influenced by environmental factors such as humidity, UV exposure, preservation methods, and sample age (Nsubuga et al., 2004; Panasci et al., 2011). Thus, studies that use fecal samples where DNA is likely to be degraded may result in genotyping errors such as allelic dropout and false alleles (Fernando and Melnick, 2001). To reduce genotyping error, multi-tube approaches are recommended, where consensus results are derived from up to seven independent PCR reactions per sample (Taberlet et al., 1996). Moreover, a two-step nested PCR is recommended for genotyping errors that are due to a limited amount of DNA (Imaizumi et al, 2014).
The use of communal latrine sites is reported in otters (Barocas et al., 2016). Therefore, the sexing of fecal DNA samples will be more informative when it is used in conjunction with identifying individuals (Park et al., 2011). Thus, the method described in our study could be used in parallel with genotyping for individual identification (Murphy et al., 2021).
CONCLUSION
In conclusion, both SRY and ZFX/ZFY markers are reliable markers for identifying sex in both Asian small-clawed and smooth-coated otters, when used with good quality samples such as tissue and blood. However, meticulous DNA extraction and PCR techniques should be practiced, especially when dealing with non-invasive samples such as otter spraints. These sexing methods may also be useful in wildlife forensics and demographic studies. Future studies employing fecal samples collected from captive otters of known sex will help to further examine the robustness of SRY and ZFX/ZFY markers for non-invasive samples. Moreover, the current sex markers can be used in conjunction with microsatellite markers for simultaneous analysis for sex and individual identification.
Acknowledgements: We thank the Department of Wildlife and National Parks (DWNP) Peninsular Malaysia for providing us the tissue samples and permit (Permit no.: W-00042-15-20 & W-00528-16-21) for our study. We would like to thank Haizan for his assistance with otter spraint collection.
REFERENCES
Aasen, E., Medrano, J.F. (1990). Amplification of the ZFY and ZFX genes for sex identification in humans, cattle, sheep and goats. Bio/technology (Nature Publishing Company), 8: 1279–1281. https://doi.org/10.1038/nbt1290-1279
Abdul-Patah, P., Nur-Syuhada, N., Md-Nor, S., Sasaki, H., Md-Zain, B.M. (2014). Habitat and food resources of otters (Mustelidae) in Peninsular Malaysia. AIP Conference Proceedings, 1614: 693–699. https://doi.org/10.1063/1.4895286
Barocas, A., Golden, H.N., Harrington, M.W., McDonald, D.B., Ben-David, M. (2016). Coastal latrine sites as social information hubs and drivers of river otter fission-fusion dynamics. Animal Behaviour, 120: 103-114. http://dx.doi.org/10.1016/j.anbehav.2016.07.016
Ben-David, M., Blundell, G.M., Kern, J.W., Maier, J.A.K., Brown, E.D. and Jewett, S.C. (2005). Communication in River Otters: Creation of Variable Resource Sheds for Terrestrial Communities. Ecology, 86: 1331-1345. https://doi.org/10.1890/04-0783
Burhanuddin, H.M.N., Ahmad, N. (1990). A survey on the distribution of otters in Pulau Pinang and Perlis. Journal of Wildlife and Parks, 9: 53-58. http://wildlife.gov.my/images/document/penerbitan/jurnal/Jil91990.pdf
Dallas, J.F., Carss, D.N., Freda, M., Koepfli, K.-P., Kruuk, H., Piertney, S.B., Bacon, P.J. (2000). Sex identification of the Eurasian otter Lutra lutra by PCR typing of spraints. Conservation Genetics, 1: 181–183. https://doi.org/10.1023/A
DeMay, S.M., Becker, P.A., Rachlow, J.L., Waits, L.P. (2017). Genetic monitoring of an endangered species recovery: Demographic and genetic trends for reintroduced pygmy rabbits (Brachylagus idahoensis). Journal of Mammalogy, 98: 350-364. https://doi.org/10.1093/jmammal/gyw197
Dos Remedios, N., Lee, P.L.M., Székely, T., Dawson, D.A., Küpper, C. (2010). Molecular sex-typing in shorebirds: a review of an essential method for research in evolution, ecology and conservation. Wader Study Group Bulletin, 117: 109–118. https://www.waderstudygroup.org/article/2208/
Estes, J.A., Palmisano, J.F. (1974). Sea otters: their role in structuring nearshore communities. Science, 185: 1058–1060. https://doi.org/10.1126/science.185.4156.1058
Fechner P.Y. (1996). The role of SRY in mammalian sex determination. Acta paediatrica Japonica: Overseas edition, 38: 380–389. https://doi.org/10.1111/j.1442-200x.1996.tb03512.x
Fernando, P.J., Melnick, D.J. (2001). Molecular sexing eutherian mammals. Molecular Ecology Notes, 1: 350–353. https://doi.org/10.1046/j.1471-8278.2001.00112.x
Fisher, R.A. (1930). The Genetical Theory of Natural Selection. Clarendon Press, Oxford. http://dx.doi.org/10.5962/bhl.title.27468
Forgacs, D., Wallen, R. L., Boedeker, A. L., & Derr, J. N. (2019). Evaluation of fecal samples as a valid source of DNA by comparing paired blood and fecal samples from American bison (Bison bison). BMC genetics, 20: 22. https://doi.org/10.1186/s12863-019-0722-3
Hattori, K., Burdin, A.M., Onuma, M., Suzuki, M., Ohtaishi, N. (2003). Sex determination in the sea otter (Enhydra lutris) from tissue and dental pulp using PCR amplification. Canadian Journal of Zoology, 81: 52–56. https://doi.org/10.1139/z02-219
Hernández-Romero, P.C., Guerrero, J.A., Valdespino, C. (2015). Morphological variability of the cranium of Lontra longicaudis (Carnivora: Mustelidae): a morphometric and geographic analysis. Zoological Studies, 54: 1-12. 10.1186/s40555-015-0127-6
Hon, N., Neak, P., Khov, V., Cheat, V. (2010). Food and habitat of Asian small-clawed otters In Northeastern Cambodia. IUCN Otter Specialist Group Bulletin, 27: 12 – 23. https://www.iucnosgbull.org/Volume27/Hon_et_al_2010.html
Hrovatin, K., Kunej, T. (2018). Genetic sex determination assays in 53 mammalian species: Literature analysis and guidelines for reporting standardization. Ecology and Evolution, 8: 1009–1018. https://doi.org/10.1002/ece3.3707
Hussain, S.A., Gupta, S.K. and de Silva, P.K. (2011). Biology and Ecology of Asian Small-Clawed Otter Aonyx cinereus (Illiger, 1815): A Review. IUCN Otter Spec. Group Bull. 28 (2): 63 – 75 https://www.iucnosgbull.org/Volume28/Hussain_et_al_2011.html
Imaizumi, K., Taniguchi, K., Ogawa, Y. (2014). DNA survival and physical and histological properties of heat-induced alterations in burnt bones. International Journal of Legal Medicine, 128: 439–446. https://doi.org/10.1007/s00414-014-0988-y
Ishigami, J, Ambu, LN, Tuuga, A and Tsubouchi, T (2017). The Second Recent Record of Hairy-Nosed Otter (Lutra sumatrana) in Sabah, Malaysia. IUCN Otter Spec. Group Bull. 34 (2): 67 – 72 https://www.iucnosgbull.org/Volume34/Ishigami_et_al_2017.html
Khan, M.S., Dimri, N.K., Nawab, A., Ilyas, O., Gautam, P. (2014). Habitat use and conservation status of smooth-coated otters Lutrogale perspicillata in Upper Gangetic Basin, India. Animal Biodiversity and Conservation, 37: 69–76. https://doi.org/10.32800/abc.2014.37.0069
Khoo, M., Basak, S., Sivasothi, N., de Silva, P.K., Reza Lubis, I. (2021). Lutrogale perspicillata. The IUCN Red List of Threatened Species 2021: e.T12427A164579961. https://dx.doi.org/10.2305/IUCN.UK.2021-3.RLTS.T12427A164579961.en
Kruuk, H., Kanchanasaka, B., O’Sullivan, S., Wanghongsa, S. (1994). Niche separation in three sympatric otters Lutra perspicillata, L. lutra and Aonyx cinerea in Huai Kha Khaeng, Thailand. Biological Conservation, 69: 115–120. https://doi.org/10.1016/0006-3207(94)90334-4
Lande, R. (1993). Risks of population extinction from demographic and environmental stochasticity and random catastrophes. The American Naturalist, 142: 911–927. http://www.jstor.org/stable/2462690
Lim, Q.L., Tan, Y.L., Ng, W.L., Yong, C., Ismail, A., Rovie-Ryan, J.J., Rosli, N., Annavi, G. (2020). Molecular sexing and preliminary assessment of population sex ratio of the endangered Malayan tapir (Tapirus indicus) in Peninsular Malaysia. Scientific reports, 10(1), 3973. https://doi.org/10.1038/s41598-020-60552-y
Melisch, R., Kusumawardhani, L., Asmoro, P.B., Lubis, I.R. (1996). The otters of West Java: a survey of their distribution and habitat use and a strategy towards a species conservation programme. PHPA/Wetlands International – Indonesia Programme, Bogor, Indonesia.
Mucci, N., Randi, E. (2007). Sex identification of Eurasian otter (Lutra lutra) non-invasive DNA samples using ZFX/ZFY sequences. Conservation Genetics, 8: 1479–1482. https://doi.org/10.1007/s10592-007-9303-5
Murakami, M., Fujise, H., Lee, Y.S., Matsuba, C., Fujitani, H. (2001). Reliable sex identification of dogs by modified PCR/RFLP analysis. Journal of Veterinary Medical Science, 63: 679-681. https://doi.org/10.1292/jvms.63.679
Murphy, M.A., Waits, L.P., Kendall, K.C. (2003). The influence of diet on faecal DNA amplification and sex identification in brown bears (Ursus arctos). Molecular Ecology, 12: 2261-2265. https://doi.org/10.1046/j.1365-294X.2003.01863.x
Murphy, S.M., Adams, J.R., Waits, L.P., Cox, J.J. (2021). Evaluating otter reintroduction outcomes using genetic spatial capture–recapture modified for dendritic networks. Ecology and Evolution, 11: 15047–15061. https://doi.org/10.1002/ece3.8187
Nichols, J.D., Armstrong, D.P. (2012). Monitoring for reintroductions. In J. G. Ewen, D.P. Armstrong, K.A. Parker, P.J. Seddon (Eds.), Reintroduction biology: Integrating science and management, pp. 223– 255. Wiley- Blackwell. https://doi.org/10.1002/9781444355833.ch7
Nunney, L. (1993). The influence of mating system and overlapping generations on effective population size. Evolution, 47: 1329–1341. https://doi.org/10.2307/2410151
Nsubuga, A.M., Robbins, M.M., Roeder, A.D., Morin, P.A., Boesch, C., Vigilant, L. (2004). Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Molecular ecology, 13: 2089–2094. https://doi.org/10.1111/j.1365-294X.2004.02207.x
Pages, M., Maudet, C., Bellemain, E., Taberlet, P., Hughes, S., Hanni, C. (2009). A system for sex determination from degraded DNA: A useful tool for palaeogenetics and conservation genetics of ursids. Conservation Genetics, 10: 897–907. https://doi.org/10.1007/s10592-008-9650-x
Pain, D ((2020). A Review of the Hairy-Nosed Otter (Lutra sumatrana) in Borneo and a Recent Sighting at Danum Valley Conservation Area, Sabah. IUCN Otter Spec. Group Bull. 37 (3): 171- 178 https://www.iucnosgbull.org/Volume37/Pain_2020.html
Palsboll, P.J., Vader, A., Bakke, I., and El-Gewely, M.R. (1992). Determination of sex in cetaceans by the polymerase chain reaction. Canadian Journal of Zoology, 70: 2166-2170. https://doi.org/10.1139/z92-292
Panasci, M., Ballard, W.B., Breck, S., Rodriguez, D., Densmore, L.D., III, Wester, D.B. and Baker, R.J. (2011). Evaluation of fecal DNA preservation techniques and effects of sample age and diet on genotyping success. The Journal of Wildlife Management, 75: 1616-1624. https://doi.org/10.1002/jwmg.221
Park, H.C., Han, T.Y., Kim, D.C., Min, M.S., Han, S.Y., Kim, K.S., Lee, H. (2011). Individual identification and sex determination of Eurasian otters (Lutra lutra) in Daegu city based on genetic analysis of otter spraint. Genes and Genomics, 33: 653–657. https://doi.org/10.1007/s13258-011-0051-z
Pilgrim, K.L., Mckelvey, K.S., Riddle, A.E., Schwartz, M.K. (2005). Felid sex identification based on noninvasive genetic samples. Molecular Ecology Notes, 5: 60-61. https://doi.org/-10.1111/j.1471-8286.2004.00831.x
Ryazanov, D.A., Maminov, M.K. (1996). Age and sex determination from canine teeth in sea otter. Zoologicheskii Zhurnal, 75: 593-601.
Salahshour, F (2016). Confirmed Sighting of Lutra sumatrana in the Ulu Muda Forest Reserve in Kedah, Malaysia. IUCN Otter Spec. Group Bull. 33 (1): 68 – 72 https://iucnosgbull.org/Volume33/Salahshour_2016.html
Sekiguchi, T., Sasaki, H., Kurihara, Y., Watanabe, S., Moriyama, D., Kurose, N., Matsuki, R., Yamazaki, K., Saeki, M. (2010). New methods for species and sex determination in three sympatric Mustelids, Mustela itatsi, Mustela sibirica and Martes melampus. Molecular ecology resources, 10: 1089–1091. https://doi.org/10.1111/j.1755-0998.2010.02842.x
Shariff, S. (1984). Some observations on otters at Kuala Gula, Perak and National Park, Pahang. Journal of Wildlife and National Parks, 3: 75-88. http://wildlife.gov.my/images/document/-penerbitan/jurnal/Jil31984.pdf
Sharma, S., Chee-Yoong, W., Kannan, A., Rama Rao, S., Abdul-Patah, P., Ratnayeke, S. (2022). Identification of three Asian otter species (Aonyx cinereus, Lutra sumatrana, and Lutrogale perspicillata) using a novel noninvasive PCR-RFLP analysis. Ecology and Evolution, 12: e9585. https://doi.org/10.1002/ece3.9585
Shaw, C.N., Wilson, P.J., White, P.N. (2003). A reliable molecular method of sex determination for mammals. Journal of Mammalogy, 84: 123–128. https://doi.org/10.1644/1545-1542(2003)084%3C0123:ARMMOG%3E2.0.CO;2
Sivasothi, N., Burhannudin, M. N. (1994). A review of otters (Carnivora: Mustelidae: Lutrinae) in Malaysia and Singapore. Hydrobiologia, 285: 151–170. https://doi.org/10.1007/BF00005663
Smith, D.A., Ralls, K., Hurt, A., Adams, B., Parker, M., Maldonado, J.E. (2006). Assessing reliability of microsatellite genotypes from kit fox faecal samples using genetic and GIS analyses. Molecular ecology, 15: 387–406. https://doi.org/10.1111/j.1365294X.2005.028-41.x
Statham, M.J., Turner, P.D., Reilly, C.O. (2007). Molecular Sex Identification in Five Mustelid Species. Zoological Studies, 46: 600–608. https://zoolstud.sinica.edu.tw/Journals/46.5/600.pdf
Taberlet, P., Camarra, J.J., Griffin, S., Uhrès, E., Hanotte, O., Waits, L.P., Dubois-Paganon, C., Burke, T., Bouvet, J. (1997). Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Molecular ecology, 6: 869–876. https://doi.org/10.1046/j.1365-294X.1997.00251.x
Taberlet, P., Griffin, S., Goossens, B., Questiau, S., Manceau, V., Escaravage, N., Waits, L. P., Bouvet, J. (1996). Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic acids research, 24: 3189–3194. https://doi.org/10.1093/nar/24.16.3189
Taberlet, P., Mattock, H., Dubois-Paganon, C., Bouvet, J. (1993). Sexing free-ranging brown bears Ursus arctos using hairs found in the field. Molecular Ecology, 2: 399-403. https://doi.org/-10.1111/j.1365-294X.1993.tb00033.x
Tan, H.H. (2015). A Roadkill Record of a Hairy-Nosed Otter (Lutra sumatrana) from Selangor, Peninsular Malaysia IUCN Otter Spec. Group Bull. 32 (1): 8 – 11 https://www.iucnosgbull.org/Volume32/Tan_2015.html
Theng, M., Sivasothi, N., Tan, H.H. (2016). Diet of the smooth-coated otter Lutrogale perspicillata (Geoffroy, 1826) at natural and modified sites in Singapore. Raffles Bulletin of Zoology, 64: 290–301. http://zoobank.org/uurn:lsid:zoobank.org:pub:A06B968E-5237-4F63-AF88019-CF7AC5581
Trinca, C.S., Eizirik, E. (2012). Molecular sexing of Neotropical otter (Lontra longicaudis) noninvasive samples. Conservation Genetics Resources, 4: 575–577. https://doi.org/10.1007-/s12686-011-9595-0
Weston, M.A., Kraaijeveld-Smit, F.J.L., McIntosh, R., Sofronidis, G., Elgar, M.A. (2004). A male-biased sex-ratio in non-breeding Hooded Plovers on a salt-lake in Western Australia. Pacific Conservation Biology, 9: 273–277. https://www.publish.csiro.au/PC/PC040273
Wildlife Conservation Act, 2010, Malaysia (Act No. 716). An Act to provide for the protection and conservation of wildlife and for matters connected therewith. https://storage.unitedwebnetwork.com/files/478/2bcd898fbf196a7cc36b99572fbc3a70.pdf
Wilson, KF and Namaskari, N (2020). Analysis of the Food-Web of a Population of Smooth-Coated Otters Lutrogale perspicillata (Mammalia: Mustelidae) in a Saline Littoral Mangrove Habitat. IUCN Otter Spec. Group Bull. 37 (1): 3 – 19 https://www.iucnosgbull.org/Volume37/Wilson_Namaskari_2020.html
Wright, L., de Silva, P.K., Chan, B., Reza Lubis, I., Basak, S. (2021). Aonyx cinereus. The IUCN Red List of Threatened Species 2021: e.T44166A164580923. https://dx.doi.org/10.2305/-IUCN.UK.2021-3.RLTS.T44166A164580923.en
Yoxon, P., Yoxon, K. (2014). Estimating Otter Numbers Using Spraints: Is It Possible? Journal of Marine Biology, 2014: 1-6. https://doi.org/10.1155/2014/430683
Zenke, P., Zorkóczy, O.K., Lehotzky, P., Ózsvári, L., Pádár, Z. (2022). Molecular Sexing and Species Detection of Antlered European Hunting Game for Forensic Purposes. Animals, 12: 1–14. https://doi.org/10.3390/ani12030246
Zhu, Y., Liu, H.Y., Yang, H. Q., Li, Y.D., Zhang, H.M. (2017). Factors affecting genotyping success in giant panda fecal samples. PeerJ, 5: e3358 https://peerj.com/articles/3358/


Résumé: Détermination du Sexe de la Loutre Cendrée (Aonyx cinereus) et de la Loutre à Pelage Lisse (Lutrogale perspicillata) par Analyse de l’ADN des Épreintes
L’identification du sexe dans les populations naturelles fournit des informations sur la démographie de la population, les relations entre espèces et les stratégies comportementales. Nous avons évalué l’applicabilité de deux marqueurs sexuels établis, à savoir le gène de la région déterminante du sexe (SRY) et le gène du doigt de zinc (ZFX/ZFY) sur la loutre cendrée (Aonyx cinereus) et la loutre à pelage lisse (Lutrogale perspicillata) en Malaisie. Nous avons utilisé ces amorces pour amplifier une partie des gènes SRY et ZFX/ZFY afin d’amplifier l’ADN extrait d’échantillons de tissus de loutres de sexe connu et avons ensuite testé leur efficacité pour amplifier des échantillons non invasifs avec 20 échantillons d’épreintes de loutres sauvages. Les amplicons ont ensuite été observés par résolution sur gel d’agarose. Résultats de l’amplification de l’ADN des six échantillons de tissus de loutres typées avec précision selon le sexe pour les deux marqueurs : Le marqueur SRY a produit un fragment de 70 pb qui s’amplifie uniquement chez les mâles, tandis que le marqueur ZFX/ZFY a produit un seul fragment de 180 pb chez les deux sexes. Les échantillons d’épreintes prélevés dans la nature ont produit des amplicons de taille correcte pour le marqueur SRY, mais le succès de l’amplification a été faible avec le marqueur ZFX/ZFY, où cinq des 20 échantillons ont donné un test PCR attendu dans au moins deux répétitions sur trois, cinq ont donné des bandes parasites, et aucun test PCR n'a été détecté avec le reste. Des techniques de laboratoire rigoureuses sont nécessaires lorsqu’il s’agit d’échantillons non invasifs tels que les épreintes, où les abandons alléliques et les tests PCR provenant d’ADN non ciblé constituent des problèmes courants. Pour tester davantage la robustesse des deux marqueurs sur des échantillons non invasifs, des échantillons fécaux de sexe connu prélevés sur des loutres en captivité sont recommandés pour de futures études. En conclusion, les marqueurs sexuels SRY et ZFX/ZFY ont fonctionné de manière fiable pour les loutres cendrées et à pelage lisse. Le succès des deux marqueurs sexuels suggère que cette méthode est applicable à la médecine légale de la faune et aux études démographiques sur les loutres en Malaisie et ailleurs dans leur aire de répartition.
Revenez au dessus
Resumen: Determinación del Sexo de la Nutria de Uñas Pequeñas Asiática (Aonyx cinereus) y la Nutria Lisa (Lutrogale Perspicillata) mediante Análisis de ADN en Fecas
La identificación del sexo en poblaciones naturales proporciona elementos para comprender la demografía, las relaciones de parentesco, y las estrategias de comportamiento. Evaluamos la aplicabilidad de dos marcadores sexuales establecidos, concretamente el gen de la región determinante del sexo (en inglés SRY) y el gen dedo de zinc (en inglés ZFX/ZFY) en la nutria de uñas pequeñas asiática (Aonyx cinereus) y la nutria lisa (Lutrogale perspicillata) en Malasia. Utilizamos estos primers para amplificar una porción de los genes SRY y ZFX/ZFY para amplificar el ADN extraído de las muestras de tejido de nutrias de sexo conocido y luego testeamos su eficacia para amplificar muestras no-invasivas con 20 muestras de fecas de nutrias silvestres. Los amplicons fueron luego observados resolviendo en gel de agarosa. Los resultados de la amplificación de ADN de todas las muestras de tejido (6 muestras) identificaron en forma precisa el sexo de las nutrias para ambos marcadores. El marcador SRY rindió un producto 70 bp que amplifica solamente a partir de machos, mientras que el marcador ZFX/ZFY produjo un único fragmento de 180 bp en ambos sexos. Las muestras colectadas en la naturaleza rindieron amplicones de tamaño correcto para el marcador SRY, sin embargo con el marcador ZFX/ZFY el éxito de amplificación fue bajo -cinco de las 20 muestras produjeron el producto PCR esperado en por lo menos dos de tres replicados, cinco produjeron bandas espurias, y en el resto no fueron detectados productos PCR. Se requieren técnicas de laboratorio estrictas al trabajar con muestras no-invasivas como fecas, siendo problemas comunes el dropout alélico y los productos PCR de ADN no-blanco. Para testear más profundamente la robustez de ambos marcadores en muestras no-invasivas, se recomienda para futuros estudios tomar muestras de fecas de sexo conocido colectadas de nutrias en cautiverio. En conclusión, tanto los marcadores sexuales SRY y ZFX/ZFY se desempeñaron confiablemente para las nutrias de uñas pequeñas Asiáticas y las nutrias lisas. El éxito de ambos marcadores sexuales sugiere que éste método es aplicable en estudios forenses y demográficos de nutrias en Malasia y en todo el rango de distribución de estas especies más allá de Malasia.
Vuelva a la tapa






